|
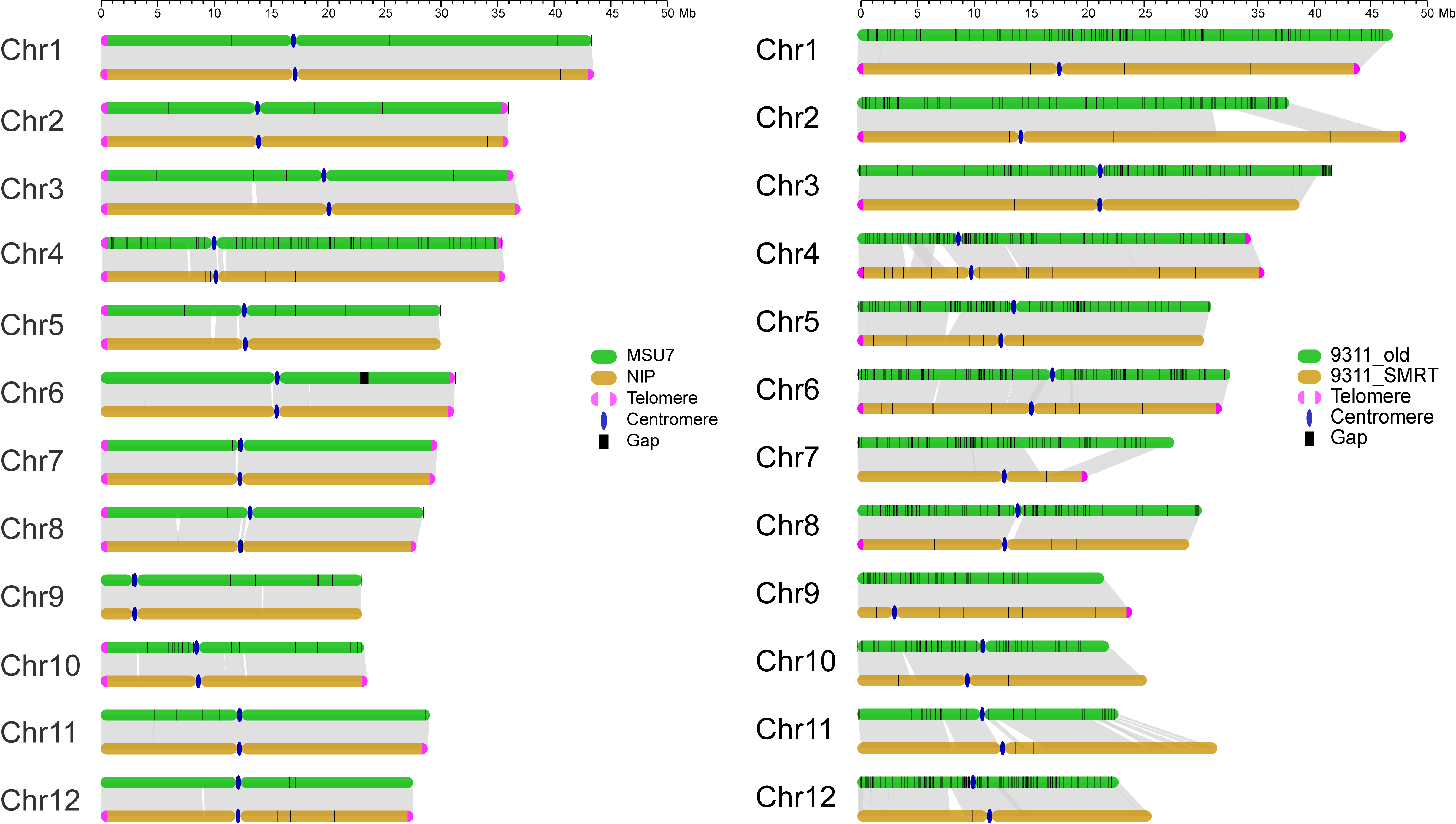
Japonica group cultivar Nip and Indica group cultivar 93-11 are the two main cultivated varieties and parental lines for breeding in Asia. This database provides a vastly improved de novo assembly and annotation of Nip and 93-11 reference genomes using SMRT-seq technique. The newly assembled Nip and 93-11 reference genomes displayed 2.2-fold (contig N50: 16,974kb versus 7,711kb) and 460-fold (9,640kb versus 21kb) higher contiguity than Nip MSU7 (http://rice.plantbiology.msu.edu/index.shtml) and 93-11 BGI (http://rise2.genomics.org.cn/page/rice/index.jsp) genomes, respectively. The new reference assembly has greatly reduced the gap number, from 905 gaps of MSU7 to 18 gaps of BRI for Nip, and 54,600 gaps of BGI to 65 gaps of BRI for 93-11. The new genome assemblies corrected 152 and 8,347 gene annotation errors in Nip and 93-11, respectively, including corrections of gene orientation and gene structure, and identified a number of new genes.
|
|
|
|
Feature |
Nip-BRI |
93-11-BRI |
|
Genome size (bp) |
380,697,840 |
395,529,940 |
Contig number |
146 |
195 |
Gap number |
18 |
65 |
Gap ratio (%) |
0.0005 |
0.0016 |
GC content (%) |
43.54 |
43.64 |
Gene number |
39,251 |
38,504 |
Gene length (bp) |
111,836,585 |
116,600,764 |
Average gene length (bp) |
2,849.27 |
3,028.28 |
CDS length (bp) |
42,283,449 |
43,045,962 |
CDS length/gene (bp) |
1,077.26 |
1,117.96 |
|
Orientation correction |
26 |
1,300 |
Structure correction |
55 |
1,318 |
Orientation & structure correction |
3 |
219 |
New genes |
68 |
5,510 |
Common genes |
38,789 |
31,458 |
|
|
|